- Ironwood Ridge High School
- Nighthawk Proteomics Institute
Manno, Theodore
Page Navigation
- About the Teacher
- Contact Info
- Bioscience I
- Bioscience II
- Bioscience Internship
- Biology (General)
- Nighthawk's Nest Biotechnology News
- Laboratory Gallery
- Classroom Gallery
- Nighthawk Chess Collective
- Nighthawk Origami & Tangram
- The Nighthawk Phylogeny Project
- Nighthawk Proteomics Institute
- Biostatistics @ The Nighthawk's Nest
- Ironwood Writer's Workshop
- The Ridgewood Methods
- Nighthawk Biotechnology Class Mascots
- Nighthawk Biotechnology In Action--Video Montage
-
Nighthawk Proteomics Institute
Information is power.
Proteomics is the large-scale study of proteins, which are vital parts of living organisms. Proteins have myriad functions, such as formation of muscle tissue structural fibers, enzymatic digestion of food, infection protection from antibodies, hormonal signaling, and DNA replication. Thus, understanding protein structure and function is key to innovation in biotechnology.
A proteome is the entire set of proteins produced or modified by an organism, and studying proteomes demands an interdisciplinary approach that generally involves acquiring genetic information. Although IRHS students learn about protein purification, mass spectrometry, and assays (e.g., using ELISA and Western Blots to test for HIV) during their time in our Bioscience program, these topics are taught as routine portions of our curriculum. To supplement their knowledge of this exciting field, student members of Nighthawk Biotechnology-Next Generation generally focus on the bioinformatical aspects of proteomics at "The Institute." In so doing, IRHS students learn the Central Dogma of biology (DNA to mRNA to protein) by starting a long-term project with the DNA of living matter which they extracted and sequenced, and ending the project 2-3 months later with a polypeptide chain and 3D structual model of a protein that corresponds to their DNA sequence.
The Nighthawk Proteomics Institute is a 3-week intensive set of lesson plans that focuses on meshing current trends in bioinformatics with widely-available proteomic-related technology by expanding upon student conclusions from The Nighthawk Phylogeny Project. Thus, Nighthawk Proteomics Institute serves as a capstone to the long-term class studies of the Central Dogma. To study phylogeny, students spend hours in our spacious, 21st-century laboratory extracting DNA from plant leaves sampled from gardens on campus, then amplify (multiply) genetic material from a particular gene in their sample using polymerase chain reaction (PCR). After cleaning their PCR product with washing buffer, centrifugation, and other procedures, students confirm the purity of their sample is suitable for Sanger Sequencing with a Thermofisher Nanodrop and send their sample to the University of Arizona Genetics Core (UAGC). Within a few days, they receive a "FASTA file," with the DNA sequence of their amplified region of interest.

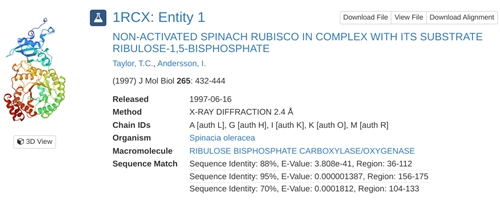
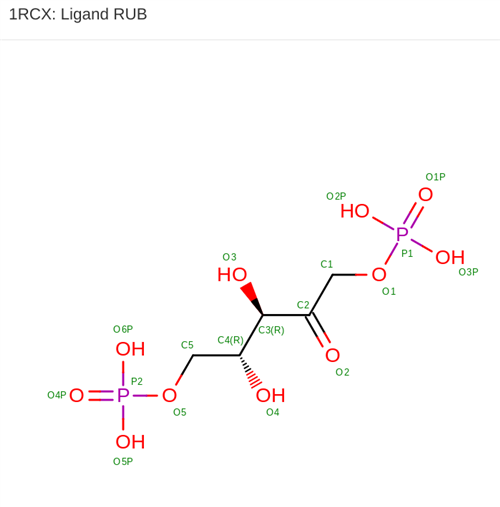
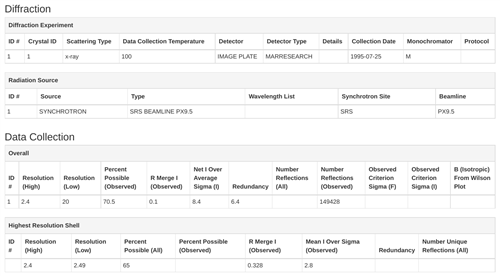
In addition to modeling phylogenies of plants following alignment of DNA sequences from the entire class with an algorithm, students search for their amino acid sequence (which they translate from their FASTA DNA sequence with 4-Peaks software) using the Protein Data Bank (PDB). This process generates assessments to confirm that the presence of a protein coded for by their DNA sequence is present in a particular organism (such as spinach).

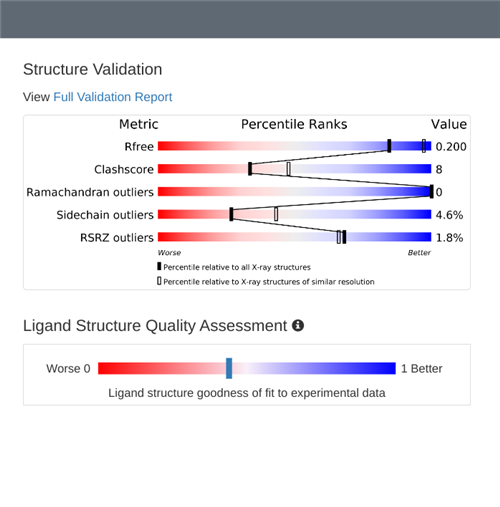
Students continue researching the protein that corresponds to the sequence from their sample, generating 3D visuals, bibliographic citations, and other information.
 ...
... ...
...
...
...
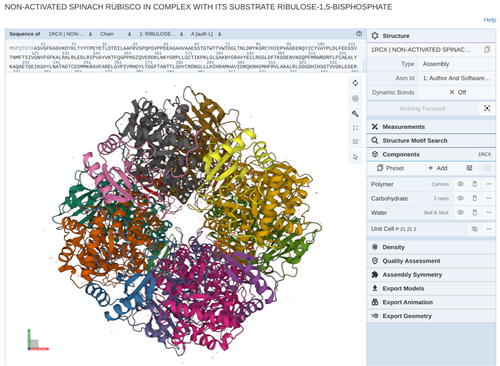
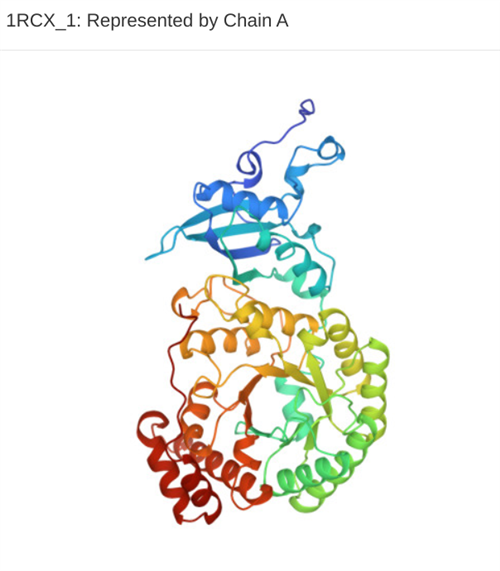
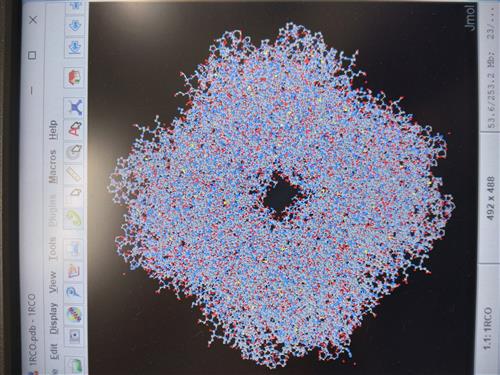
Students use other programs such as JMOL to visualize the protein related to their DNA sequence and create models of how the protein might change structure and function if certain parts of their sequence changed.
Students are rewarded with unlimited potential for future research, and with awesome stickers ("Chowl" stickers!!)
Come be a part of something special at Nighthawk Biotechnology-Next Generation.
We are the future. And the future is now.
#nighthawkbiotechnextgen

